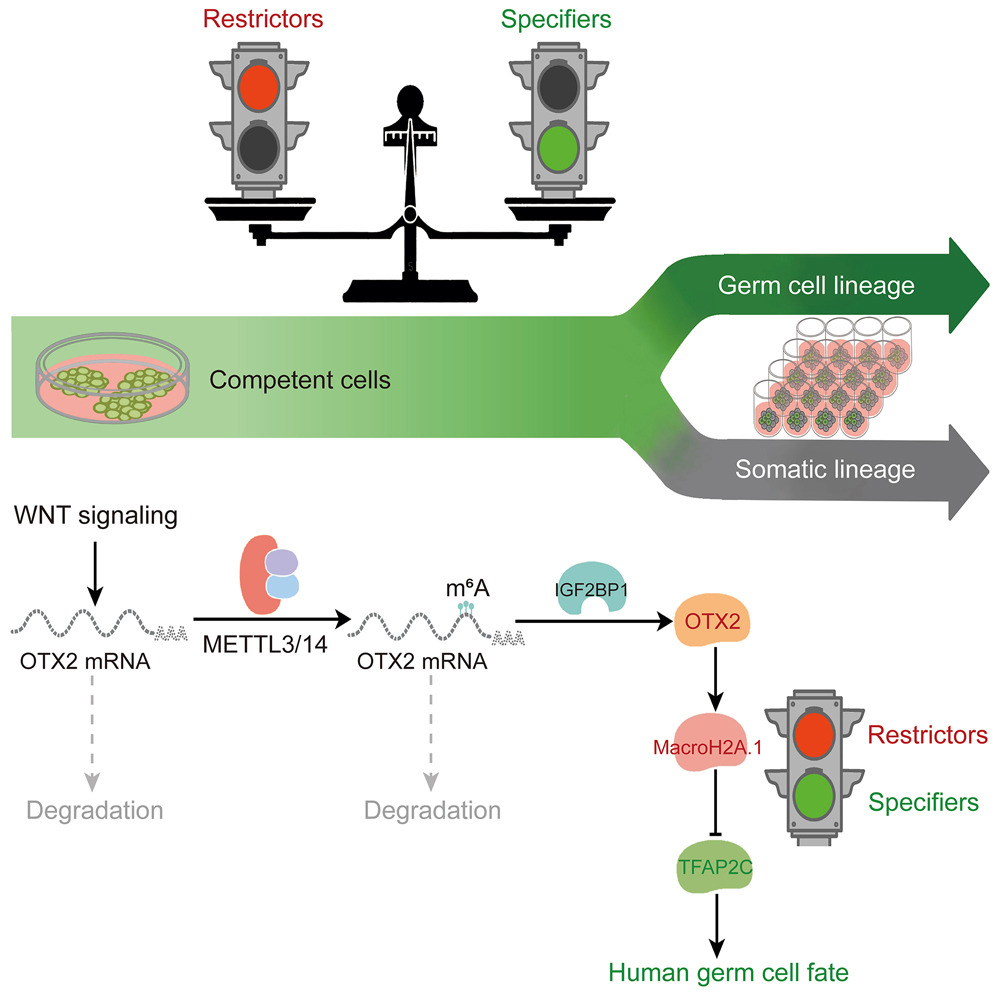
IGF2BP1 restricts the induction of human primordial germ cell fate in an m6A-dependent manner.
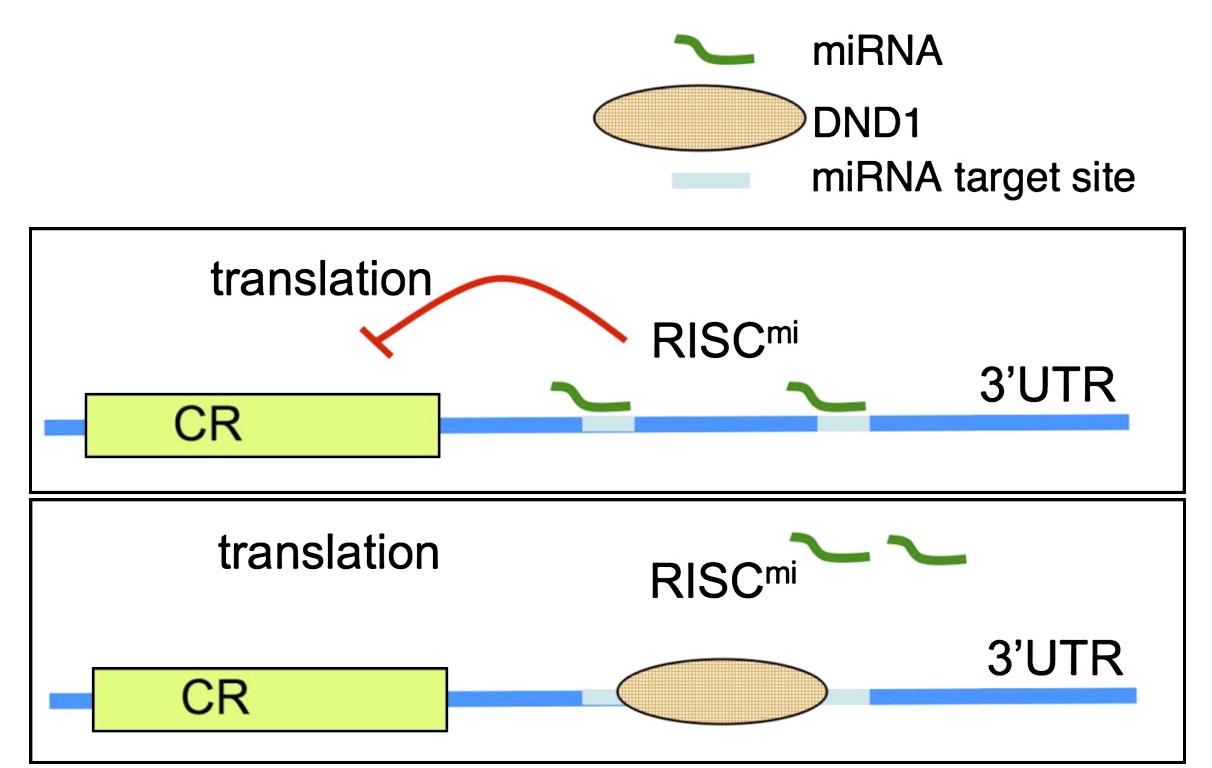
RNA-binding proteins DND1 and NANOS3 coordinately suppress the translation of SOX4 mRNAs in processing bodies for restricting the entry of germ cell lineage.
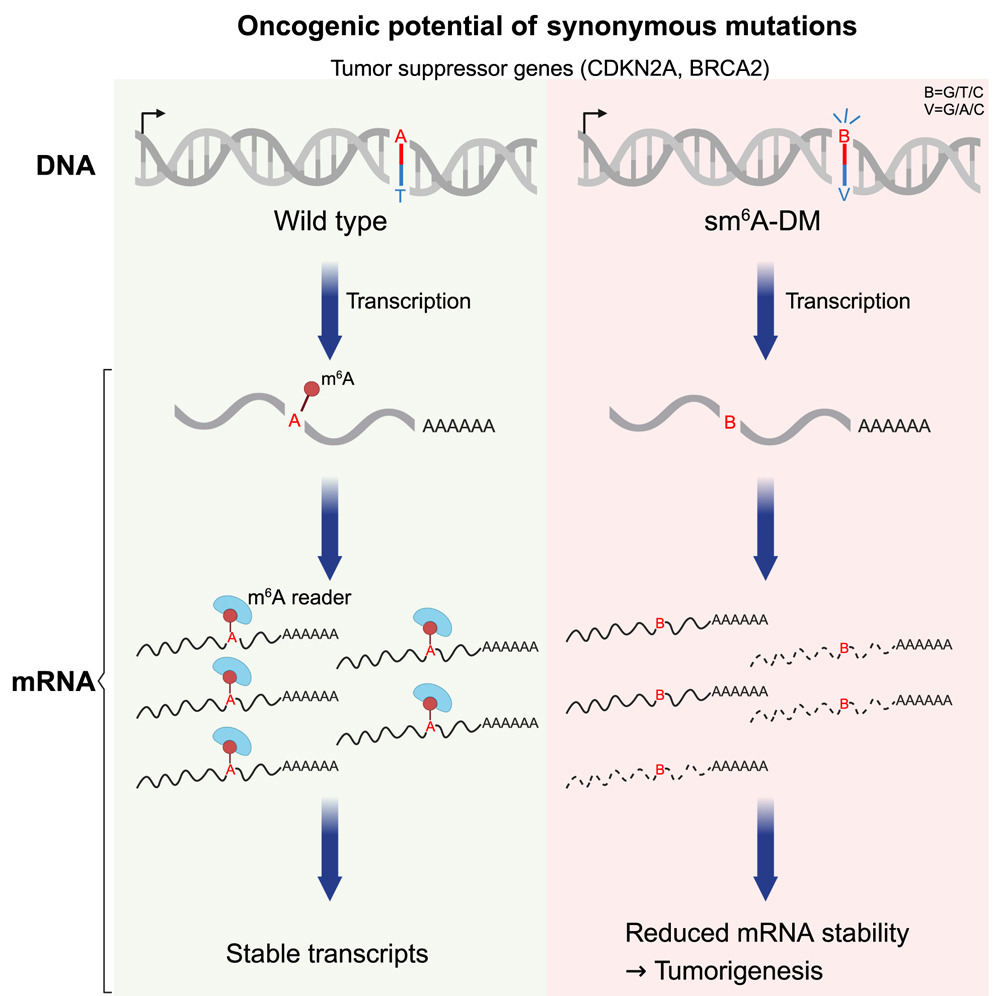
Synonymous mutations promote tumorigenesis by disrupting m6A-dependent mRNA metabolism.
44. Ma, J.*, Yu, H.*, Yao, S., Yan, Y., Gu, Z., Wang, Z., Huang, H.#, & Chen, D.# (2025). Making cells inter-connected for signaling communication: a developmental view of cytonemes. Cell communication and signaling: CCS, 23(1), 241.
https://doi.org/10.1186/s12964-025-02229-5
43. Zhang, J., Gu, Y., Tong, L., Feng, B., Dong, S., Shao, Q., Chen, Y., Tu, H., Wang, Z., Wang, Y., Li, X., Yu, H., Lin, Z., Wang, X., Li, Z., Ai, Z., Xiang, Y., Jiang, Z., Jin, Z., Li, Z., Chen, Y., Shen, Z., Huang, C., Liu, J., Liu, J., Xu, P., Yu, Y., Xia, P., Liang, H., Huang, H.#, & Chen, D.# (2025). IGF2BP1 restricts the induction of human primordial germ cell fate in an m6A-dependent manner. Cell stem cell, S1934-5909(25)00181-X. Advance online publication.
https://doi.org/10.1016/j.stem.2025.05.001
42. Feng, B.*, Chen, Y.*, Tu, H.*, Zhang, J., Tong, L., Lyu, X., Irving, A. T., & Chen, D.# (2025). Transcriptomic Analysis of the m6A Reader YTHDF2 in the Maintenance and Differentiation of Human Embryonic Stem Cells. Stem cells, sxaf032. Advance online publication.
https://doi.org/10.1093/stmcls/sxaf032
41. Wang, Z.*, Yu, H.*, Gu, Z., Shi, X., Ma, J., Shao, Q., Yao, Y., Yao, S., Xu, Y., Gu, Y., Dai, J., Liu, Q., Shi, J., Qi, R., Jin, Y., Liu, Y., Shen, X., Huang, W., Liu, H., Jin, M., Liu, W., Brook, M.# & Chen, D.# (2025). RNA-binding proteins DND1 and NANOS3 cooperatively suppress the entry of germ cell lineage. Nature communications, 16(1), 4792.
https://doi.org/10.1038/s41467-025-57490-6
40. Yu, H., & Chen, D.# (2025). In preprints: the extracellular matrix influences primordial germ cell behavior. Development, 152(9), dev204861.
https://doi.org/10.1242/dev.204861
39. Yu, H., Wang, Z., Ma, J., Wang, R., Yao, S., Gu, Z., Lin, K., Li, J., Young, R. S., Yu, Y., Yu, Y., Jin, M., & Chen, D. (2025). The establishment and regulation of human germ cell lineage. Stem cell research & therapy, 16(1), 139.
https://doi.org/10.1186/s13287-025-04171-2
38. Lan, Y., Xia, Z., Shao, Q., Lin, P., Lu, J., Xiao, X., Zheng, M., Chen, D. #, Dou, Y.#, & Xie, Q.# (2025). Synonymous mutations promote tumorigenesis by disrupting m6A-dependent mRNA metabolism. Cell, S0092-8674(25)00095-9. Advance online publication.
https://doi.org/10.1016/j.cell.2025.01.026
37. Gu, Y., Chen, J., Wang, Z., Shao, Q., Li, Z., Ye, Y., Xiao, X., Xiao, Y., Liu, W., Xie, S., Tong, L., Jiang, J., Xiao, X., Yu, Y., Jin, M., Wei, Y.#, Young, R. S.#, Hou, L.#, & Chen, D.# (2024). Integrated analysis and systematic characterization of the regulatory network for human germline development. Journal of genetics and genomics, S1673-8527(24)00306-0.
https://doi.org/10.1016/j.jgg.2024.11.005
36. Shi, X., Xi, C., Dong, B., Yan, Z., Liu, W.#, Gao, S.#, & Chen, D.# (2024). Maternal infection with SARS-CoV-2 during early pregnancy induces hypoxia at the maternal-fetal interface. Cell proliferation, e13749.
https://doi.org/10.1111/cpr.13749
35. Zhou, Z., Tong, L., Chen, Y., Wang, R., Shen, Y., & Chen, D.# (2025). Dual-Selection Strategy for Generating Knock-Out Lines of Human Embryonic Stem Cells. Journal of cellular and molecular medicine, 29(2), e70259.
https://doi.org/10.1111/jcmm.70259
34. Yuan, Z., Han, X., Xiao, M., Zhu, T., Xu, Y., Tang, Q., Lian, C., Wang, Z., Li, J., Wang, B., Li, C., Xiang, X., Jin, R., Liu, Y., Yu, X., Zhang, K., Li, S., Ray, M., Li, R., Gruzdev, A., Shao, S., Shao, F., Wang, H., Lian, W., Tang, Y., Chen, D., Lei, Y., Jin, X., Li, Q., Long, W., Huang, H., DeMayo, F., & Liu, J.# (2024). Overexpression of ELF3 in the PTEN-deficient lung epithelium promotes lung cancer development by inhibiting ferroptosis. Cell death & disease, 15(12), 897.
https://doi.org/10.1038/s41419-024-07274-5
33. Feng, X., Li, Z., Liu, Y., Chen, D., & Zhou, Z.# (2024). CRISPR/Cas9 technology for advancements in cancer immunotherapy: from uncovering regulatory mechanisms to therapeutic applications. Experimental hematology & oncology, 13(1), 102.
https://doi.org/10.1186/s40164-024-00570-y
32. Chen, Y., Zhou, Z., Chen, Y., & Chen, D.# (2024). Reading the m6A-encoded epitranscriptomic information in development and diseases. Cell & bioscience, 14(1), 124.
https://doi.org/10.1186/s13578-024-01293-7
31. Zhang, J., Tong, L., Liu, Y., Li, X., Wang, J., Lin, R., Zhou, Z., Chen, Y., Chen, Y., Liu, Y., & Chen, D.#(2024). The regulatory role of m6A modification in the maintenance and differentiation of embryonic stem cells. Genes & diseases, 11(5), 101199.
https://doi.org/10.1016/j.gendis.2023.101199
30. Wang, R., Wang, Z., Tong, L., Wang, R., Yao, S., Chen, D.#, & Hu, H.# (2024). Microfluidic Mechanoporation: Current Progress and Applications in Stem Cells. Biosensors, 14(5), 256.
https://doi.org/10.3390/bios14050256
29. Su, Y., Yu, Z., Jin, S., Ai, Z., Yuan, R., Chen, X., Xue, Z., Guo, Y., Chen, D., Liang, H., Liu, Z., & Liu, W.#(2024). Comprehensive assessment of mRNA isoform detection methods for long-read sequencing data. Nature communications, 15(1), 3972.
https://doi.org/10.1038/s41467-024-48117-3
28. Wang, X., Song, C., Ye, Y., Gu, Y., Li, X., Chen, P., Leng, D., Xiao, J., Wu, H., Xie, S., Liu, W., Zhao, Q., Chen, D., Chen, X., Wu, Q., Chen, G., & Zhang, W.# (2023). BRD9-mediated control of the TGF-β/Activin/Nodal pathway regulates self-renewal and differentiation of human embryonic stem cells and progression of cancer cells. Nucleic acids research, 51(21), 11634–11651.
https://doi.org/10.1093/nar/gkad907
27. Li, Z., Xu, H., Li, J., Xu, X., Wang, J., Wu, D., Zhang, J., Liu, J., Xue, Z., Zhan, G., Tan, B. C. P., Chen, D., Chan, Y. S., Ng, H. H., Liu, W., Hsu, C. H., Zhang, D., Shen, Y., & Liang, H.# (2023). Selective binding of retrotransposons by ZFP352 facilitates the timely dissolution of totipotency network. Nature communications, 14(1), 3646.
https://doi.org/10.1038/s41467-023-39344-1
26. Watanabe, M., Buth, J.E., Haney, J.R., Vishlaghi, N., Turcios, F., Elahi, L.S., Gu, W., Pearson, C.A., Kurdian, A,, Baliaouri, N.V., Collier, A.J., Miranda, O.A., Dunn, N., Chen, D., Sabri, S., Torre-Ubieta, L., Clark, A.T., Plath, K., Christofk, H.R., Kornblum, H.I., Gandal, M.J., & Novitch, B.G. (2022). TGFβ superfamily signaling regulates the state of human stem cell pluripotency and capacity to create well-structured telencephalic organoids. Stem cell reports, 17(10), 2220–2238.
https://doi.org/10.1016/j.stemcr.2022.08.013
25. Ai, Z., Xiang, X., Xiang, Y., Szczerbinska, I., Qian, Y., Xu, X., Ma, C., Su, Y., Gao, B., Shen, H., Bin Ramli, M. N., Chen, D., Liu, Y., Hao, J. J., Ng, H. H., Zhang, D., Chan, Y. S., Liu, W., & Liang, H.# (2022). Krüppel-like factor 5 rewires NANOG regulatory network to activate human naive pluripotency specific LTR7Ys and promote naive pluripotency. Cell reports, 40(8), 111240.
https://doi.org/10.1016/j.celrep.2022.111240
24. Jin, S., Xue, Z., Zhang, J., Wang, Z., Zhang, J., Chen, D.#, Liu, W.#, & Lin, J.# (2021). Identification of SRSF3 target mRNAs using inducible TRIBE. Biochemical and biophysical research communications, 578, 21–27.
https://doi.org/10.1016/j.bbrc.2021.09.019
23. Hancock, G. V., Liu, W., Peretz, L., Chen, D., Gell, J. J., Collier, A. J., Zamudio, J. R., Plath, K., & Clark, A. T.# (2021). Divergent roles for KLF4 and TFCP2L1 in naive ground state pluripotency and human primordial germ cell development. Stem cell research, 55, 102493.
https://doi.org/10.1016/j.scr.2021.102493
22. Chitiashvili, T., Dror, I., Kim, R., Hsu, F. M., Chaudhari, R., Pandolfi, E., Chen, D., Liebscher, S., Schenke-Layland, K., Plath, K., & Clark, A.# (2020). Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nature cell biology, 22(12), 1436–1446.
https://doi.org/10.1038/s41556-020-00607-4
21. Liu, X., Ouyang, J.F., Rossello, F.J., Tan, J.P., Davidson, K.C., Valdes, D.S., Schröder, J., Sun, Y.B..Y, Chen, J., Knaupp, A.S., Sun, G., Chy, H.S., Huang, Z., Pflueger, J., Firas, J., Tano, V., Buckberry, S., Paynter, J.M., Larcombe, M.R., Poppe, D., Choo, X.Y., O’Brien, C.M., Pastor, W.A., Chen, D., Leichter, A.L., Naeem, H., Tripathi, P., Das, P.P., Grubman, A., Powell, D.R., Laslett, A.L., David, L., Nilsson, S.K., Clark, A.T., Lister, R., Nefzger, C.M., Martelotto, L.G., Rackham, O.J.L., & Polo, J.M.# (2020). Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature, 586(7827), 101–107.
https://doi.org/10.1038/s41586-020-2734-6
20. Chen, D.*, Sun, N*., Hou, L.*, Kim, R., Faith, J., Aslanyan, M., Tao, Y., Zheng, Y., Fu, J., Liu, W., Kellis, M., & Clark, A.# (2019). Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell reports, 29(13), 4568–4582.e5.
https://doi.org/10.1016/j.celrep.2019.11.083
19. Chen, D.*, Liu, W.*, Zimmerman, J., Pastor, W., Kim, R., Hosohama, L., Ho, J., Aslanyan, M., Gell, J., Jacobsen, S., & Clark, A.# (2018). The TFAP2C-regulated OCT4 naïve enhancer is involved in human germline formation. Cell reports, 25(13), 3591–3602.e5.
https://doi.org/10.1016/j.celrep.2018.12.011
18. Sosa, E., Chen, D., Rojas, E., Hennebold, J., Peters, K., Wu, Z., Lam, T., Mitchell, J., Tailor, R., Meistrich, M., Orwig, K., Shetty, G., & Clark, A.# (2018). Differentiation of primate primordial germ cell-like cells following transplantation into the adult gonadal niche. Nature communications, 9(1), 5339.
https://doi.org/10.1038/s41467-018-07740-7
17. Hancock, G. & Chen, D.# (2018). Another step closer to unlocking specification of primordial germ cells. AME Med Journal, 3:64.
16. Pastor, W., Liu, W., Chen, D., Ho, J., Kim, R., Hunt, T., Lukianchikov, A., Liu, X., Polo, J., Jacobsen, S., & Clark, A. (2018). TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nature cell biology, 20(5), 553–564.
https://doi.org/10.1038/s41556-018-0089-0
15. Gell, J., Zhao, J., Chen, D., Hunt, T., & Clark, A.# (2018). PRDM14 is expressed in germ cell tumors with constitutive overexpression altering human germline differentiation and proliferation. Stem cell research, 27, 46–56.
https://doi.org/10.1016/j.scr.2017.12.016
14. Chen, D. & Clark, A.# (2018). Mitochondrial DNA selection in human germ cells. Nature cell biology, 20(2), 118–120.
https://doi.org/10.1038/s41556-017-0029-4
13. Chen, D., Liu, W., Lukianchikov, A., Hancock, G., Zimmerman, J., Lowe, M., Kim, R., Galic, Z., Irie, N., Surani, A., Pastor, W., Ho, J., Jacobsen, S., & Clark, A.# (2017). Germline competency of human embryonic stem cells depends on EOMESODERMIN. Biology of reproduction, 97(6), 850–861.
https://doi.org/10.1093/biolre/iox138
12. Chen, D.# & Hancock, G. (2017). Crosslinking and immunoprecipitation: a new route for dead end. AME Medical Journal 2:119.
11. Clark, A. T.#, Gkountela, S., Chen, D., Liu, W., Sosa, E., Sukhwani, M., Hennebold, H., & Orwig, K. (2017). Primate primordial germ cells acquire transplantation potential by Carnegie stage 23. Stem cell reports, 9(1), 329–341.
https://doi.org/10.1016/j.stemcr.2017.05.002
10. Chen, D., Gell, J. J., Tao, Y., Sosa, E., & Clark, A. T.# (2017). Modeling human infertility with pluripotent stem cells. Stem cell research, 21, 187–192.
https://doi.org/10.1016/j.scr.2017.04.005
9. Tang, Y., Geng, Q.#, Chen, D., Zhao, S., Liu, X., & Wang, Z.# (2017). Germline proliferation is regulated by somatic endocytic genes via JNK and BMP signaling in Drosophila. Genetics, 206(1), 189–197.
https://doi.org/10.1534/genetics.116.196535
8. O’Brien, C.M., Chy, H.S., Zhou, Q., Blumenfeld, S., Lambshead, J.W., Liu, X., Kie, J., Capaldo, B.D., Chung, T.L., Adams, T.E., Phan, T., Bentley, J.D., McKinstry, W.J., Oliva, K., McMurrick, P.J., Wang, Y.C., Rossello, F.J., Lindeman, G.J., Chen, D., Jarde, T., Clark, A.T., Abud, H.E., Visvader, J.E., Nefzger, C.M., Polo, J.M., Loring, J.F., & Laslett, A.L.# (2017). New Monoclonal Antibodies to Defined Cell Surface Proteins on Human Pluripotent Stem Cells. Stem cells, 35(3), 626–640.
https://doi.org/10.1002/stem.2558
7. Shan, L., Wu, C., Chen, D., Hou, L., Li, X., Wang, L., Chu, X., Hou, Y., & Wang, Z.# (2017). Regulators of alternative polyadenylation operate at the transition from mitosis to meiosis. Journal of genetics and genomics, 44(2), 95–106.
https://doi.org/10.1016/j.jgg.2016.12.007
6. Pastor, W.A.*, Chen D.*, Liu, W.*, Kim R., Sahakyan, A., Lukianchikov, A., Plath, K., Jacobsen, S.E. & Clark, A.T.# (2016). Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell stem cell, 18(3), 323–329.
https://doi.org/10.1016/j.stem.2016.01.019
5. Chen, D., & Clark, A. T.# (2015). Human germline differentiation charts a new course. The EMBO journal, 34(8), 975–977.
https://doi.org/10.15252/embj.201591447
4. Chen, D.*, Wu, C.*, Zhao, S., Geng, Q., Gao, Y., Li, X., Zhang, Y., & Wang, Z.# (2014). Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila. PLoS genetics, 10(11), e1004797.
https://doi.org/10.1371/journal.pgen.1004797
3. Zhao, S., Chen, D., Geng, Q., & Wang, Z.# (2013). The highly conserved LAMMER/CLK2 protein kinases prevent germ cell overproliferation in Drosophila. Developmental biology, 376(2), 163–170.
https://doi.org/10.1016/j.ydbio.2013.01.023
2. Liu. Z., Huang, Y., Chen, D., & Zhang, Y.# (2011). Drosophila Acyl-CoA synthetase long-chain family member 4 regulates axonal transport of synaptic vesicles and is required for synaptic development and transmission. The Journal of neuroscience,31(6), 2052–2063.
https://doi.org/10.1523/JNEUROSCI.3278-10.2011
1. Zhang, Y., Chen, D., & Wang, Z.# (2009). Analyses of mental dysfunction-related ACSl4 in Drosophila reveal its requirement for Dpp/BMP production and visual wiring in the brain. Human molecular genetics, 18(20), 3894–3905.
https://doi.org/10.1093/hmg/ddp332